Abstract
We have generated a next-generation whole-exome sequencing data set of 2628 participants of the population-based Rotterdam Study cohort, comprising 669 737 single-nucleotide variants and 24 019 short insertions and deletions. Because of broad and deep longitudinal phenotyping of the Rotterdam Study, this data set permits extensive interpretation of genetic variants on a range of clinically relevant outcomes, and is accessible as a control data set. We show that next-generation sequencing data sets yield a large degree of population-specific variants, which are not captured by other available large sequencing efforts, being ExAC, ESP, 1000G, UK10K, GoNL and DECODE.
Similar content being viewed by others
Introduction
In the era of next-generation sequencing (NGS), the use of large population data sets to approximate variant frequencies in control populations has become common practice. The first large population-scale sequencing data set was generated by the 1000 Genomes Project,1 where an integrated genome-wide map of genetic variation was established for 2504 individuals of European, American, African and Asian descent. Another approach was made by the NHLBI ‘Grand Opportunity’ Exome Sequencing Project, in which a set of 6500 European and African Americans samples was exome sequenced.2 The recent Exome Aggregation Consortium (ExAC) is now combining exome sequencing data sets from over 60 000 unrelated individuals from different origins.3 From these large sequencing projects, it became apparent that many variants are population-specific.3 Therefore, several initiatives have generated more local data sets. The UK10K project4 contains 4000 genomes from the UK, along with 6000 exomes from individuals with selected extreme phenotypes. A collection of 3000 Finnish exomes, showed that the Finnish population had more loss-of-function variants and gene knock-outs than non-Finish Europeans.5 GoNL,6 the Dutch reference genome project, provided a local genetic map based on whole-genome sequencing of 250 Dutch trios.7 Another local data set is based on full genomes from 2636 Icelanders.8 Due to Iceland being an isolated population, deleterious variants could reach higher frequencies than in other populations. These initiatives emphasize the importance of local genetic maps to interpret clinical relevance of a potential disease-causing mutation, and indicate the differences in available population data sets that should be considered when these are used in research or clinical practice.
Within the Rotterdam Study cohort, a prospective population-based cohort study on individuals 45 years and older to investigate determinants of disease and disability in the Dutch population,9 we have generated a set of 2628 exomes for integrative genetic studies of diverse phenotypes and to serve as a local reference panel for clinical sequencing efforts.
Materials and methods
DNA samples were obtained from the Rotterdam Study, which is a prospective population-based cohort study established in 1990 studying the determinants of disease and disability in Dutch elderly individuals.9 Out of 5984 eligible participants from the RS-I cohort − based on the availability of height, weight, GWAS data and informed consent − 3284 subjects were randomly selected, as shown in Figure 1. Baseline characteristics are provided in Supplementary Table 1.
Overview of sample selection and quality control. Out of 5984 eligible samples, a final random set of 2628 exomes was generated. QC, quality control; SNP, single-nucleotide polymorphism; het/hom ratio, ratio between heterozygous and homozygous positions; Ti/Tv ratio, ratio between transitions and transversions.
Genomic DNA was prepared from whole blood and processed using the Illumina TruSeq DNA Library preparation (Illumina, Inc., San Diego, CA, USA), followed by exome capture using the Nimblegen SeqCap EZ V2 kit (Roche Nimblegen, Inc., Madison, WI, USA). Paired-end 2 × 100 bp sequencing was performed at six samples per lane on Illumina HiSeq2000 sequencer using Illumina TruSeq V3 chemistry.
Reads were demultiplexed and aligned to the human reference genome hg19 (UCSC, Genome Reference Consortium GRCh37) using the Burrows-Wheeler alignment tool (BWA version 0.7.3a10). After indel realignment and base quality score recalibration using the Genome Analysis ToolKit (GATK version 2.7.411) and masking of duplicates (Picard Tools version 1.9012), gvcf files were generated using HaplotypeCaller v3.1.1 (GATK) and genotyped using GenotypeGVCFs v3.1.1 (GATK).11 Raw genotype data was QC-ed and filtered as described in the Supplementary Information. All coding variants used in analysis are available on the European Variation Archive (http://www.ebi.ac.uk/eva/) under accession number PRJEB20726.
All detected variants were annotated based on RefSeq annotation (NCBI Reference Sequence Database) using ANNOVAR (version 2014-07-1413). The presence and allele frequencies of these variants in various databases: 1000G (v3),1 ESP (v2),2 ExAC (v0.3),3 UK10K (v1407),4 DECODE (v1501)8 and the Genome of the Netherlands (v4)6 were obtained and compared to our data set.
Results
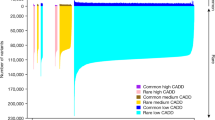
Two thousand six hundred and twenty eight samples passed technical and genetic quality control and were included in the data set (Figure 1), with an average mean depth of coverage of 55x (range 20x to 185x, median coverage of 53x). A total of 669 737 single-nucleotide variants (SNVs) and 24 019 short insertions or deletions (indels) were detected, this data set was denoted Rotterdam Study Exome Sequencing set 2 (RSX2). Of all 669 737 SNVs detected in our RSX2 data set, 439 633 (66%) were exonic. Of these, 120 677 (27.4%) were not detected in any other public database (ExAC2.0, ESP6500, 1000G, UK10K, DECODE and GoNL), as shown in Figure 2. Most of these variants (120 179; 99.6%) were found at a minor allele frequency (MAF) below 1% in our data set, 65 324 were singletons (54%) and 19 870 were doubletons (17%). The largest overlap with a single data set was with ExAC2.0 (71% of 439 633 SNVs), followed in descending order by ESP6500 (46%), 1000G (36%), UK10K (34%), GoNL (26%) and DECODE (22%).
Overlap of RSX2 with other publically available data sets. Overlap was based on only RefSeq coding SNVs which were detected in at least 1 individual in RSX2 (439 633 SNVs total). The numbers in the Venn diagrams display the number of overlapping SNVs in thousands, the numbers between parenthesis are those SNVs with MAF below 1% (386 341 total). A total of 318 586 SNVs were present in any of the 6 databases (72%). Each individual database yielded a smaller overlap, ranging from 311 017 (Exac, 71%) to 113 627 (GoNL, 26%). Almost all SNVs unique to RSX2 have a MAF<1% in the RSX2 data set (120 547; 99.6%).
Discussion
From 439 633 detected coding variants, 120 179 were absent from all six other population databases. A portion of this absence can be attributed to various biological (ie, ethnical backgrounds, isolated populations or case-series) and technical (whole-genome sequencing, exome capturing or filtering strategies and sequencing depth) differences, the remainder is most likely due to population-specific variance.
The smallest overlap with DECODE is partly due to the lower sequencing depth and stronger filtering strategy in that data set, resulting in fewer variants in general. In addition, the genetically isolated status of the Icelandic population warrants fewer genetic variability and smaller overlap with RSX2.8 Despite originating from a similar population, the small overlap with the GoNL database is likely due to its small sample size, reducing power to detect rare variants.6 A larger overlap with UK10K was observed as a result of its large sample size and related population. The differences with the UK10K data set are largely due to population-specific differences and, the selection of individuals with extreme phenotype in UK10K.4 The 1000G data set holds many more variants than RSX2, probably caused by whole-genome sequencing coverage on coding regions inaccessible by whole-exome sequencing, and by the presence of non-Caucasian individuals.1 Similarly, difference in populations and sample size leads to the ESP6500 data set to be larger than RSX2, although the selection for various case-populations might also be of influence.2 Finally, the greatest data set of ExAC2.0 contains most variants, as a result of much larger sample size and the inclusion of many different populations.3
Each data set present in this comparison contained variants not present in any of the other data sets. These results suggest that, for example, when filtering or interpreting genetic variants in a WES analysis of a Mendelian disease pedigree, both smaller population-specific data sets (such as, RSX2, GoNL, UK10K and/or DECODE) as well as large aggregation data sets (such as, ExAC) contribute information and should be used jointly to filter. Additionally, each database contributes variants not seen elsewhere, suggesting that as many databases as eligible should be considered in these types of analyses. When WES data sets are to be used as controls (eg, in a case control comparison) note should be taken that some data sets such as UK10K, ESP and ExAC2.0, contain large collections of case-series2, 3, 4 and will not provide a good representation of DNA sequence variants of any allele frequency spectrum in the normal population. Given their design and collection strategy, population-based data sets such as RSX2, DECODE and GoNL, might be better suited for this purpose, depending on the diseases and traits studies and their estimated prevalence in these databases.
References
Genomes Project C Genomes Project C, Abecasis GR Genomes Project C, Altshuler D Genomes Project C, Auton A Genomes Project C, Brooks LD Genomes Project C, Durbin RM et al: A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S et al: Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012; 337: 64–69.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T et al: Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291.
UK10K WTSI, Hinxton, UK. Available at: http://www.uk10k.org [june-2015].
Lim ET, Wurtz P, Havulinna AS, Palta P, Tukiainen T, Rehnstrom K et al: Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014; 10: e1004494.
Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A et al: The Genome of the Netherlands: design, and project goals. Eur J Hum Genet 2014; 22: 221–227.
Genome of the Netherlands C: Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014; 46: 818–825.
Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A et al: Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 2015; 47: 435–444.
Hofman A, Brusselle GG, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A et al: The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol 2015; 30: 661–708.
Li H, Durbin R : Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Wang K, Li M, Hakonarson H : ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
A Supplementary video accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
van Rooij, J., Jhamai, M., Arp, P. et al. Population-specific genetic variation in large sequencing data sets: why more data is still better. Eur J Hum Genet 25, 1173–1175 (2017). https://doi.org/10.1038/ejhg.2017.110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.110
This article is cited by
-
Chinese genetic variation database of inborn errors of metabolism: a systematic review of published variants in 13 genes
Orphanet Journal of Rare Diseases (2023)
-
In-silico mining to glean SNPs of pharmaco-clinical importance: an investigation with reference to the Indian populated SNPs
In Silico Pharmacology (2023)
-
Objectives, design and main findings until 2020 from the Rotterdam Study
European Journal of Epidemiology (2020)