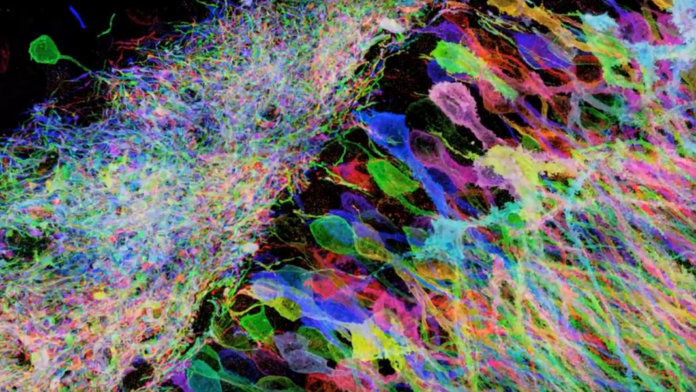
Like the fiber optic cables that connect landline phones, the long branches of brain cells transmit information from one neuron to the next.
Unlike phone cables that can only passively carry electrical signals, however, neuronal branches are dotted with synapses—active computational powerhouses that constantly restructure themselves to form our thoughts and memories.
For a neuroscientist eager to study neural circuits and brain function, dendrites—the input cables—are a promising place to start. But with an average width tens or hundreds of times smaller than a human hair, dendrites are hard to observe. At the nanoscale level, synapses are even further beyond reach. Sure, microscopes can help by optically enlarging the tissue, but even the most state-of-the-art equipment is limited in resolution, and CSI-style “zoom and enhance” hardly ever works.
What if, instead of optically blowing up the brain, we could make it physically larger?
Sound impossible? I thought so too. But this week, a team of neuroengineers led by Dr. Edward Boyden at MIT achieved just that. By embedding the brain into a gel that swells up when pumped with water, the team blew up mouse brain tissue to roughly 20 times its original size, while preserving the normal structure and connections of neurons and their dendrites.
Using this method, aptly dubbed expansion microscopy (ExM), the team reconstructed a tiny piece of the mouse brain in 3D. Normally, dendrites entangle into a jumbled mess, making it hard to tease apart individual synaptic connections with a conventional light microscope.
With ExM, the scientists easily peeked into the dendrites’ nooks and crannies, allowing an unprecedented look at little mushroom-shaped protrusions called dendritic spines, where synapses sit.
Even wilder, the method also exposed individual protein clusters inside the spines to support normal synapse function and help create neuronal circuits.
“ExM can be used to explore neural connectivity in 3D with spatial precision sufficient for resolving individual synaptic connections,” the authors say.
“If you could reconstruct a complete brain circuit, maybe you could make a computational model of how it generates complex phenomena like decisions and emotions,” says Boyden, “you could potentially model the dynamics of the brain.”
Brain cartographers
Brain mapping has been the center of “big neuroscience” for the past few years. Ambitious billion-dollar moonshots such as the BRAIN Initiative and Europe’s Blue Brain Project all strive to develop new methods that allow neuroscientists to reconstruct in minute detail the mouse, and ultimately the human, brain.
These projects stem from a guiding principle in biology: form begets function. To understand and recreate an electrical system, the first step is to pull out its wiring blueprint. Similarly, scientists believe that mapping the physical and functional connections between neurons is our best bet at cracking the brain’s basic workings—its “neural code.”
There have certainly been a few high-profile wins in the brain-mapping domain. Just last month, an international team unveiled an automated procedure that uncovered a surprisingly large neuron that surrounds the entire mouse brain.
But all brain-charting efforts are limited by microscope resolution. Although physicists have long worked out which parameters are most crucial for overcoming these limits, improving the power and quality of microscopes has been an uphill battle.
Alice in Wonderland
Boyden’s team takes the opposite approach: rather than wrestling with light scattering and physics, why not enlarge the biological specimen?
Back in 2015, his team first demonstrated that blowing up brain tissue (without actually blowing it up into bits) is possible. The key is using brains that have been preserved in formalin, cutting it up into very thin sections and injecting each section with a dense, evenly generated hyper-absorbent gel normally used in baby diapers.
Before expansion, the team tagged the proteins they wanted to image—for example, proteins at the synapse—with a custom molecule called an antibody. On one end, the antibody contains a chemical group that binds to the polymers that make up the expandable gel, which anchors the tagged proteins to the gel. On the other end is a small single strand of DNA that acts like a bar code.
After tagging, the tissue is treated with a gentle meat tenderizer to melt away proteins that normally hold it together. The team then plopped the brain-gel into water, and watched it expand to roughly four and a half times its normal size, allowing the tags to drift away from each other like dots on an inflating balloon.
This enlarged brain slice is then ready for imaging with a common type of microscope called a confocal microscope. To visualize the tagged proteins, the researchers added different colored fluorescent probes that bind to the DNA bar codes—in this way, one type of protein is labeled yellow; another green; another blue, and so on.
The method allowed the scientists to achieve a resolution of roughly 60 nanometers, about ten times larger than without treating the tissue.
Yet Boyden was unsatisfied.
“Individual biomolecules are much smaller than that, say five nanometers or even smaller,” he says. But expanding the tissue even more made it unstable and destroyed normal brain structure.
Through the looking glass
To get around the growth limit, the team modified their approach so that after the first expansion, the brain-gel was once again embedded into a new gel that blows up the tissue a second time—something they call “iterative expansion” (iExM).
“iExM typically resulted in expansion ratios of ~4.5–5.5× in the first round and ~4× in the second round for a total increase of ~16–22×,” the authors explain.
And the difference showed. Focusing on the mouse hippocampus, a brain region important for learning and memory, researchers could clearly distinguish the sharp outlines of the dendrites and their spines. However, when it came to the proteins that make up the synapses, they showed up as blurry little blobs.
In contrast, synaptic proteins were sharply labeled in tissue treated with the new iExM. Not only could the researchers see “signature” proteins that lie on each side of the synapse, they could also directly visualize the neurochemical receptors that sit on synapses. These receptors allow information to jump from one neuron to the next by acting as part of the machinery that transforms electrical signals to chemical ones.
In this way, researchers may be able to map the unique receptor configurations that switch neural circuits on and off.
“iExM may be useful for analyzing the varying nanoscale configurations of proteins across brain circuits and regions, because it can support large-volume imaging with nanoscale precision,” the authors say.
And indeed they went large. Expanding a brainbow mouse, in which different neurons are randomly labeled with different colors, the team zoomed in on a normally densely-packed region of the hippocampus.
These spines are normally hard to resolve without expansion, they explain, but with iExM the “number, size, position and shapes of dendritic spines were easily visualized.”
A sharper future
The team is now gearing up for a third round of expansion, which hypothetically would enable a resolution of 5 nanometers. The roadblock is finding sufficiently small fluorescent probes that can squeeze together to latch onto two nearby proteins before expanding. Alternatively, scientists could also develop new probes that tag their protein after expansion.
While getting sharper images is great, the ultimate goal is being able to trace the wiring of complete brain circuits, says Boyden.
He already has an idea: temporal multiplexing. Currently, only a few colored probes can be used to image proteins in a sample—once and done.
Temporal multiplexing essentially turns the tissue into multi-use. Researchers can tag a group of proteins as usual, image the tissue, then strip away the probes and re-attach new ones.
This allows an exponential number of proteins to be probed with each round, the researchers say.
“By combining iterative expansion with temporal multiplexing, we could in principle have essentially infinite-color, nanoscale-resolution imaging over large 3-D volumes,” says Boyden. “Things are getting really exciting now that these different technologies may soon connect with each other.”
Image Credit: Ed Boyden/YouTube